A focus of the Beatty Group at OHSU is developing new chemical tools for illuminating human diseases. We have been inspired to use innovative, cross-disciplinary approaches to identify and investigate the molecular basis of human diseases, including tuberculosis (TB) and breast cancer. Since 2012, we have worked on the following research projects:
1. Chemical probes for mycobacterial pathogens.
2. VIP Tags: Genetic tags for correlative light and electron microscopy.
3. Imaging the HER signaling network in breast cancer.
4. Synthesis of new fluorescent and fluorogenic probes.
1. Chemical tools for detecting hydrolase activities in Mycobacterium tuberculosis (Mtb).
Beatty group team members: Katie Tallman (PMCB), Dr. Samantha Levine, Hailey Dearing, and Kaylyn Devlin
Collaborators: Prof. Gyanu Lamichhane, Prof. Aaron Wright, and Dr. Vivian Lin.
Project funding: NIH (NIAID: R01 AI149737), OHSU School of Medicine, Collins Medical Trust, Medical Research Foundation of Oregon
The human pathogen Mtb is the causative agent of tuberculosis (TB). TB is the deadliest disease in human history, and kills nearly 2 million people every year. There are ~10 million new cases a year and an estimated one-third of humans harbor Mtb in a dormant state. These asymptomatic, latent infections impede TB eradication due to the long-term potential for reactivation. Moreover, three million cases of TB go undiagnosed each year, which contributes substantially to the spread of the disease. At OHSU, our research team is using chemistry to accelerate discovery of TB biomarkers and drug targets by analyzing the function of enzymes implicated in latent and active TB.
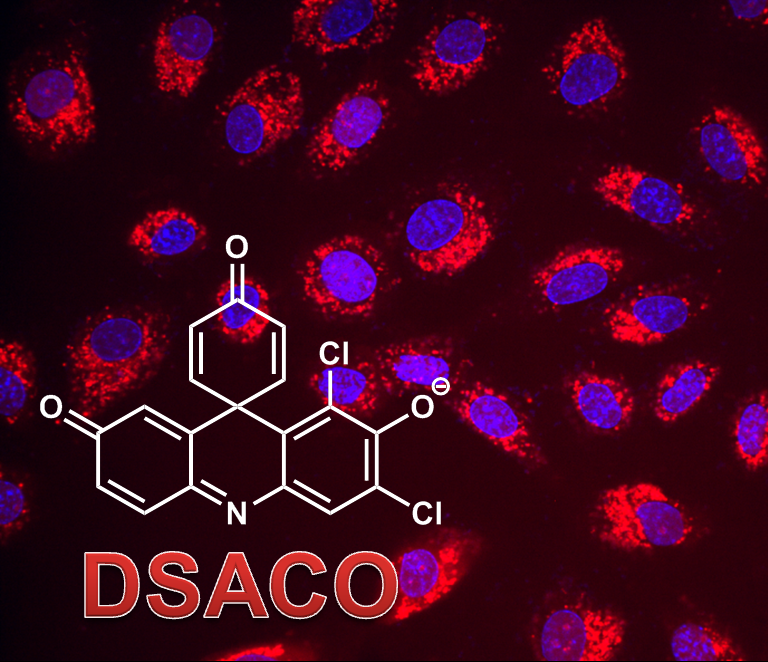
Our strategy has been to use chemistry to accelerate discoveries. We use activity-based probes to highlight functional enzymes in mycobacterial cells or lysates. This approach circumvents the need for purified enzymes or genetically-modified pathogens. Dr. Beatty initiated this research program as a postdoc (UC Berkeley, Bertozzi Lab), synthesizing a fluorogenic probe for sulfatases. This probe revealed that there are differences in sulfatase activity patterns (fingerprints) between different mycobacterial species and strains.
At OHSU, we developed a new set of probes for investigating hydrolase regulation in M. tuberculosis. In 2014, we reported two fluorogenic probes for esterase and lipase detection. These probes were used to identify active esterases in gel-resolved lysates, an assay platform developed in the Beatty Lab. Next we targeted mycobacterial lipases. Those and other probes were used to identify esterases in active, dormant, and reactivating cultures. In summary, we found that there is dynamic regulation of Mtb esterases in different stages of growth. This makes esterases good targets for classifying Mtb infections. In the future, such probes could be used to characterize lesions in the lungs of TB patients.
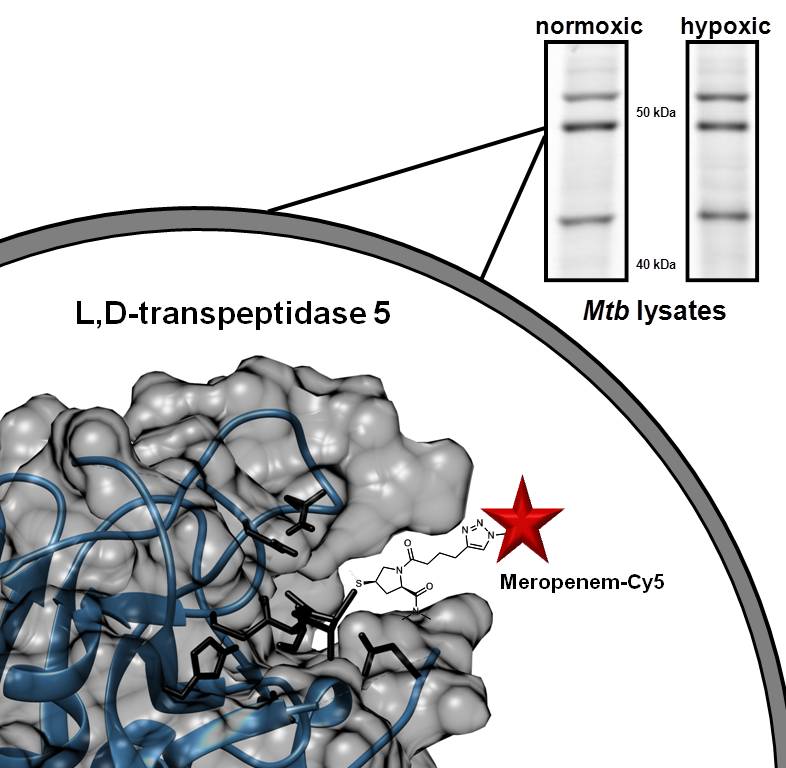
In the last few years, we have shifted our focus towards understanding drug-resistance in TB. We are focusing on establishing the the efficacy of drugs targeting mycobacterial cell wall biosynthesis. For example, we are identifying the targets of beta-lactam antibiotics. While beta-lactams, such as penicillin, are rarely used to treat TB today there have been some exciting reports that beta-lactams (e.g., meropenem-clavulanate) can cure drug-resistant TB. In 2021, we published a set of probes ideal for identifying beta-lactam drug targets in Mtb. Our current work is focused on analyzing the regulation of two classes of enzymes: PBPs and LDTs. These are great targets for drugs because both classes of enzyme maintain the structure and rigidity of the bacterial cell wall. With the support of NIADID, we are using a novel probe (meropenem-biotin) combined with chemo-proteomics to identify drug targets in dormant and replicating Mtb.
We are fortunate to have a team of collaborators on this project, including Prof. Gyanu Lamichhane (JHU), Prof. Aaron Wright (Baylor), and Dr. Vivian Lin (PNNL).
2. VIP Tags: Genetic tags for correlative light and electron microscopy.
Beatty group team members: Julia Doh (PMCB), Dr. Miguel Macias-Contreras, Ms. Savannah Tobin, Dr. Hannah Zane, Dr. Jon White, Alexa Suyama, and Kaylyn Devlin
Collaborators: Caroline Enns, Claudia Lopez, Dan Zuckerman, Ujwal Shinde, and Young Hwan Chang
Project funding: NIH (NIGMS: R01GM122854)
Recent advances in imaging instrumentation and computational analysis have created new opportunities for investigating the molecular basis of diseases with remarkable detail. It is now possible to interrogate features ranging in size from angstroms to centimeters, which enables investigations into tissue architectures, neuronal connections, organelle coordination, signaling networks, and molecular organization. These are representative examples of the types of studies that would immediately benefit from a versatile technology for labeling and tracking proteins across size scales. We foresee an increased reliance on multi-color, multi-scale microscopy for investigating proteins associated with human diseases. The central obstacle that has decelerated progress in this area is the shortage of methods for labeling proteins for multi-scale microscopy.
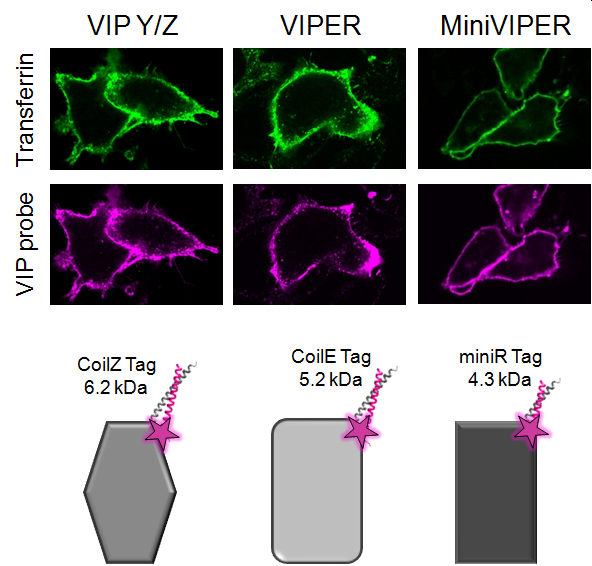
To address this unmet need, we created a new concept for labeling proteins with reporters compatible with multi-scale microscopy. Versatile interacting peptide (VIP) tags were designed for analyzing proteins across size scales and different imaging systems. Tags can be used in fixed or living cells, enabling dynamic studies of receptors. A key innovation of this technology was creating tags that enable protein labeling with reporters matched to the application, such as quantum dots (Qdots) for correlative light and EM (CLEM). VIP tags are small (<6 kDa), making them less likely than other tags (e.g., 30 kDa GFP) to disrupt normal protein function. Labeling is achieved via a heterodimeric coiled-coil interaction between a tag and a reporter-conjugated probe peptide. Labeling is robust and specific in cells, making VIP tags an excellent alternative to immunolabeling.
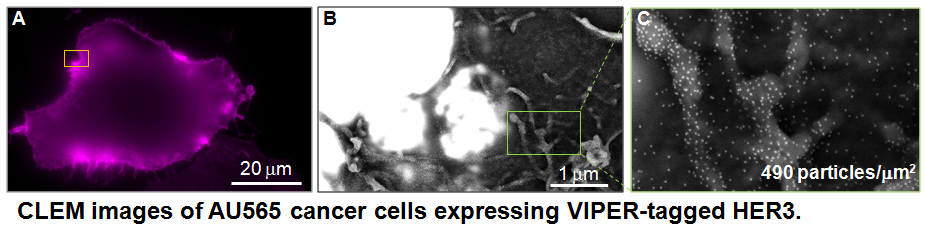
Our strategy combines genetically-encoded peptide tags with a palette of reporter chemistries for labeling cellular nanostructures. In 2017, we published our first demonstration of using VIP tags to label cellular proteins (ChemBioChem, 2017). Those VIP tags were small, target-specific, easy to use, and compatible with diverse chemical reporters, including bright organic fluorophores (fluorescein, rhodamine, and AF647) and quantum dots. The reporter chemistry can be selected and optimized for different applications, which makes this technology a powerful resource for imaging studies. Next we described VIPER, a tag formed by dimerization between the genetically-encoded tag (CoilE) and a probe peptide (CoilR). Using confocal FM, we showed that VIPER can highlight organelles or track receptor endocytosis in live cells by “pulse-chase” labeling and time-lapse imaging. We used VIPER to image TfR1-mediated iron uptake by CLEM. Our CLEM studies included quantitative image analysis, demonstrating that VIPER labeling was more efficient than immunolabeling. In 2020, we described MiniVIPER, a 4.3 kDa tag for imaging cellular proteins and for translocating proteins inside cells.
In ongoing work, we are developing and validating additional VIP tags. In unpublished work (2022), we are creating a set of bioorthogonal tags for observing up to three targets at once in cells. Long-term, this technology will enable novel investigations into the spatial organization of cells during differentiation and migration, in response to drug treatment or infection, as well as a variety of other scenarios. Furthermore, we will use VIP tags to add novel functionality to proteins. This project benefits from a creative team of forward-thinking chemists and collaborators willing to try new technologies.
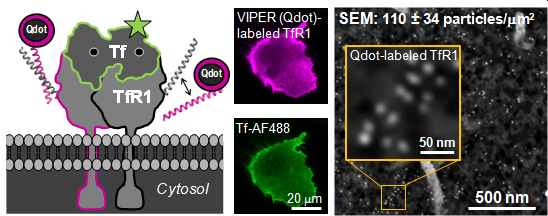
3. Imaging the HER signaling network in breast cancer.
Beatty group team members: Julia Doh and Dr. Kaylyn Devlin
Collaborators: Tiera Liby, Dr. Jim Korkola and Dr. Joe Gray
Project funding: Women’s Circle of Giving and the Knight Cancer Institute
We are collaborating with Dr. Joe Gray and Dr. Jim Korkola to investigate mechanisms of drug resistance in breast cancer. HER2 amplification occurs in ~25% of all breast cancer and is associated with poor prognosis. HER2-targeted therapeutics are clinically available, but their efficacy in patients has not lived up to pre-clinical promise. Multiple reasons for resistance to HER2-targeted inhibitors have been identified, including reactivation of HER2 signaling through feedback mechanisms. HER2 is thought to require HER3 as a partner for oncogenic signaling, as knockout of HER3 phenocopies HER2 knockout in HER2-amplified cells. We are using VIP tags to capture multi-color views of the dynamics of HER2-HER3 signaling.
4. Synthesis of new fluorescent and fluorogenic probes.
Beatty group team members: Dr. Samantha Levine, Katie Tallman, and Nick Lopez
Many of the most widely used reporter chemistries are fluorescent or fluorogenic (e.g., “turn-on”) probes. For imaging within living systems, the most useful fluorophores excite and emit between 600 and 800 nm. This region is biologically “quiet”, with little endogenous absorption, scattering, or autofluorescence. However, there are few far-red chemical reporters. We are using our expertise in color chemistry to develop new fluorescent and fluorogenic probes. Some of our research in this area used the far-red fluorophore DDAO (Proc. Natl. Acad. Sci. USA 2013, ACS Chem. Biol. 2016, ACS Infect. Dis. 2016, ChemBioChem 2014). We have also synthesized far-red carbazines that can be converted into enzyme-activated fluorophores (Chem. Commun., 2016). In the future, we team will continue to develop novel molecular imaging agents for biomedical research.